pacman::p_load(tidyverse,rio)
stroke <- import("dataset/stroke.csv")5 数据清洗
前面,我们花了不少篇幅,做了一些基础性的操作。在正式进入数据可视化和分析以前,我们还要学会如何将导入的原始数据,通过某些操作 把数据转化成可以直接分析的整洁数据,这个过程就是数据清洗。英文名有很多,data cleaning,data wrangling,data manipulation,data munging, data transformation, data processing等
5.1 清洗步骤
这些步骤,前三步,顺序在某些情况下可以变化,比如可以先数据变形
- 列操作
- 行操作
- 数据变形
- 数据归一
- 清除重复和空值
- 数据合并
5.2 列-行操作
想一想,拿到表格数据后,我们应该做些什么。首先就是先留下自己想要的,不需要的内容我们可以剔除,比如,病人的姓名是隐私,一般不需要(剔除列);一些记录不符合纳入标准,需要剔除掉,比如患者年龄太小或太大(剔除行)。这就需要列-行的操作。先进行列操作,是我的习惯,你当然也可以先操作行。
需要加载的包,需要读取的数据如下
在进行各种操作以前,我们一般先整体上看一下这个数据的状况,我们可以用一个函数glimpse()
glimpse(stroke)Rows: 213
Columns: 12
$ doa <chr> "17/2/2011", "20/3/2011", "9/4/2011", "12/4/2011", "12/4…
$ dod <chr> "18/2/2011", "21/3/2011", "10/4/2011", "13/4/2011", "13/…
$ status <chr> "alive", "alive", "dead", "dead", "alive", "dead", "aliv…
$ sex <chr> "male", "male", "female", "male", "female", "female", "m…
$ dm <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes",…
$ gcs <int> 15, 15, 11, 3, 15, 3, 11, 15, 6, 15, 15, 4, 4, 10, 12, 1…
$ sbp <int> 151, 196, 126, 170, 103, 91, 171, 106, 170, 123, 144, 23…
$ dbp <int> 73, 123, 78, 103, 62, 55, 80, 67, 90, 83, 89, 120, 120, …
$ wbc <dbl> 12.5, 8.1, 15.3, 13.9, 14.7, 14.2, 8.7, 5.5, 10.5, 7.2, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ stroke_type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "hospital", …该数据集的变量名解释
- doa : date of admission
- dod : date of discharge
- status : event at discharge (alive or dead)
- sex : male or female
- dm : diabetes (yes or no)
- gcs : Glasgow Coma Scale (value from 3 to 15)
- sbp : Systolic blood pressure (mmHg)
- dbp : Diastolic blood pressure (mmHg)
- wbc : Total white cell count
- time2 : days in ward
- stroke_type : stroke type (Ischaemic stroke or Haemorrhagic stroke)
- referral_from : patient was referred from a hospital or not from a hospital
5.2.1 列[选择-重命名-新列]
列-行操作,用到的包主要是dplyr,隶属于tidyverse系列,按照习惯,我们直接加载tidyverse即可(前面已经加载过)。
5.2.1.1 选择列select()
通过列号或者列名选择
stroke2 <- stroke %>%
select(1,3,sex,dm,stroke_type) %>%
glimpse()Rows: 213
Columns: 5
$ doa <chr> "17/2/2011", "20/3/2011", "9/4/2011", "12/4/2011", "12/4/2…
$ status <chr> "alive", "alive", "dead", "dead", "alive", "dead", "alive"…
$ sex <chr> "male", "male", "female", "male", "female", "female", "mal…
$ dm <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes", "…
$ stroke_type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "IS"…有时候,我们希望大部分列都保留,只剔除不想要的列,只需在列号或列名前加-
stroke3 <- stroke %>%
select(-1,-sex) %>%
glimpse()Rows: 213
Columns: 10
$ dod <chr> "18/2/2011", "21/3/2011", "10/4/2011", "13/4/2011", "13/…
$ status <chr> "alive", "alive", "dead", "dead", "alive", "dead", "aliv…
$ dm <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes",…
$ gcs <int> 15, 15, 11, 3, 15, 3, 11, 15, 6, 15, 15, 4, 4, 10, 12, 1…
$ sbp <int> 151, 196, 126, 170, 103, 91, 171, 106, 170, 123, 144, 23…
$ dbp <int> 73, 123, 78, 103, 62, 55, 80, 67, 90, 83, 89, 120, 120, …
$ wbc <dbl> 12.5, 8.1, 15.3, 13.9, 14.7, 14.2, 8.7, 5.5, 10.5, 7.2, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ stroke_type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "hospital", …5.2.1.2 重命名列rename()
对列名字不太满意?
stroke3 %>%
rename(type = stroke_type, DM=dm) %>%
glimpse()Rows: 213
Columns: 10
$ dod <chr> "18/2/2011", "21/3/2011", "10/4/2011", "13/4/2011", "13/…
$ status <chr> "alive", "alive", "dead", "dead", "alive", "dead", "aliv…
$ DM <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes",…
$ gcs <int> 15, 15, 11, 3, 15, 3, 11, 15, 6, 15, 15, 4, 4, 10, 12, 1…
$ sbp <int> 151, 196, 126, 170, 103, 91, 171, 106, 170, 123, 144, 23…
$ dbp <int> 73, 123, 78, 103, 62, 55, 80, 67, 90, 83, 89, 120, 120, …
$ wbc <dbl> 12.5, 8.1, 15.3, 13.9, 14.7, 14.2, 8.7, 5.5, 10.5, 7.2, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "hospital", …其实,如果我们用select()命令,在选择行的同时,可以将重命名同时进行。
想想,这样做和rename()有什么区别。
stroke4 <- stroke %>%
select( gender = sex, DM = dm) %>%
glimpse()Rows: 213
Columns: 2
$ gender <chr> "male", "male", "female", "male", "female", "female", "male", "…
$ DM <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes", "no", …5.2.1.3 创建新列 mutate()
我们想创建一个新列,是脉压值,pulse_p = 收缩压-舒张压。可以用mutate()
stroke5 <- stroke %>%
select(-1,-sex) %>%
mutate(pulse_p = sbp - dbp) %>%
glimpse()Rows: 213
Columns: 11
$ dod <chr> "18/2/2011", "21/3/2011", "10/4/2011", "13/4/2011", "13/…
$ status <chr> "alive", "alive", "dead", "dead", "alive", "dead", "aliv…
$ dm <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes",…
$ gcs <int> 15, 15, 11, 3, 15, 3, 11, 15, 6, 15, 15, 4, 4, 10, 12, 1…
$ sbp <int> 151, 196, 126, 170, 103, 91, 171, 106, 170, 123, 144, 23…
$ dbp <int> 73, 123, 78, 103, 62, 55, 80, 67, 90, 83, 89, 120, 120, …
$ wbc <dbl> 12.5, 8.1, 15.3, 13.9, 14.7, 14.2, 8.7, 5.5, 10.5, 7.2, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ stroke_type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "hospital", …
$ pulse_p <int> 78, 73, 48, 67, 41, 36, 91, 39, 80, 40, 55, 110, 87, 79,…留个作业,如果你想创建一个新列,收缩压>140或舒张压>90为高血压,并赋值为HBP,其余赋值为norm。如何做?
提示:我们可以用ifelse这个语句,ifelse(sbp>140|dbp>90, ‘HBP’, ‘norm’)
5.2.1.4 列的数据类型转换 mutate()
stroke5 %>%
mutate(status = as.factor(status)) %>%
glimpse()Rows: 213
Columns: 11
$ dod <chr> "18/2/2011", "21/3/2011", "10/4/2011", "13/4/2011", "13/…
$ status <fct> alive, alive, dead, dead, alive, dead, alive, alive, dea…
$ dm <chr> "no", "no", "no", "no", "yes", "no", "no", "yes", "yes",…
$ gcs <int> 15, 15, 11, 3, 15, 3, 11, 15, 6, 15, 15, 4, 4, 10, 12, 1…
$ sbp <int> 151, 196, 126, 170, 103, 91, 171, 106, 170, 123, 144, 23…
$ dbp <int> 73, 123, 78, 103, 62, 55, 80, 67, 90, 83, 89, 120, 120, …
$ wbc <dbl> 12.5, 8.1, 15.3, 13.9, 14.7, 14.2, 8.7, 5.5, 10.5, 7.2, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ stroke_type <chr> "IS", "IS", "HS", "IS", "IS", "HS", "IS", "IS", "HS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "hospital", …
$ pulse_p <int> 78, 73, 48, 67, 41, 36, 91, 39, 80, 40, 55, 110, 87, 79,…5.2.2 行操作 filter()
行操作的需求一般就只有一个,把我要的记录筛选出来。
比如我们创建一个名为stroke_m_9的数据集,筛选出Glasgow Coma Scale (gcs)>9分以上的男性。
stroke_m_9 <- stroke %>%
filter(sex == "male", gcs>9) %>%
glimpse()Rows: 82
Columns: 12
$ doa <chr> "17/2/2011", "20/3/2011", "22/5/2011", "28/11/2011", "25…
$ dod <chr> "18/2/2011", "21/3/2011", "23/5/2011", "29/11/2011", "26…
$ status <chr> "alive", "alive", "alive", "dead", "alive", "alive", "al…
$ sex <chr> "male", "male", "male", "male", "male", "male", "male", …
$ dm <chr> "no", "no", "no", "no", "no", "no", "no", "no", "no", "y…
$ gcs <int> 15, 15, 11, 10, 14, 15, 15, 15, 15, 15, 15, 15, 15, 14, …
$ sbp <int> 151, 196, 171, 207, 128, 143, 161, 153, 143, 116, 132, 1…
$ dbp <int> 73, 123, 80, 128, 79, 59, 107, 61, 93, 81, 81, 72, 146, …
$ wbc <dbl> 12.5, 8.1, 8.7, 10.8, 10.3, 7.1, 9.5, 11.2, 15.6, 11.0, …
$ time2 <int> 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,…
$ stroke_type <chr> "IS", "IS", "IS", "HS", "IS", "IS", "IS", "IS", "IS", "I…
$ referral_from <chr> "non-hospital", "non-hospital", "hospital", "non-hospita…5.2.3 分组和运算
有时候我们很想对数据进行分组,并分开计算。如我们想计算男女之间的血压均值分别是多少。可以分别用到group_by()和summarize()。
stroke_sex <- stroke %>%
group_by(sex) %>%
summarise(meansbp = mean(sbp, na.rm = TRUE), # na.rm参数是剔除空值
meandbp = mean(dbp, na.rm = TRUE),
meangcs = mean(gcs, na.rm = TRUE))还可试试计算统计频数,如status这个代表是生存还是死亡。
stroke_status <- stroke %>%
group_by(sex) %>%
count(status, sort = TRUE)关于列-行操作,我们来总结一下dplyr的主要操作。
大家要学会找速查表(cheatsheet),直接在搜索引擎搜索,某包的名字+cheatsheet即可,如dplyr cheatsheet。
https://dplyr.tidyverse.org/
mutate()adds new variables that are functions of existing variablesselect()picks variables based on their names.filter()picks cases based on their values.summarise()reduces multiple values down to a single summary.arrange()changes the ordering of the rows.
5.3 数据变形 tidyr
5.3.1 长宽数据转换
Reshaping Data from Wide (Fat) to Long (Tall)
宽数据变长数据
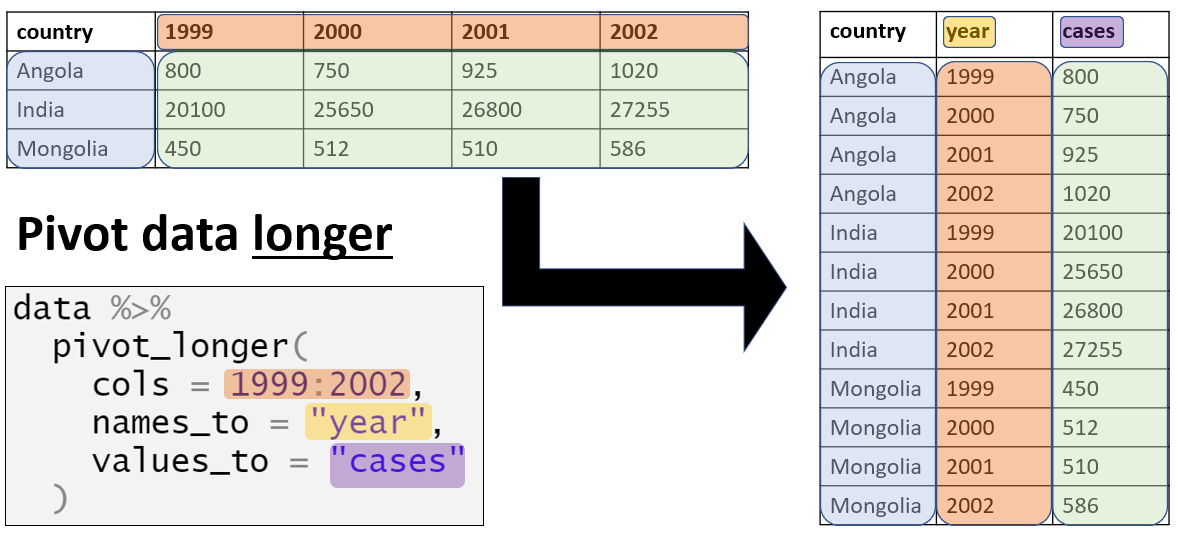
首先来看看什么是宽数据。
pwide <- read.csv("dataset/pwide.csv")
head(pwide)这个数据来自Gapminder。代表每个国家每年的民主指数。
我们看到,这个表格是把年作为了列名,但其实,我们需要的是年作为变量(variable),然后具体的年份作为数值,存入该变量。只需要一个tidyr包的pivot_longer命令即可。这样就符合整洁数据的要求。
pivot_longer(pwide,
cols = !country, #选择要改变的列
names_to = "year", #变量名合并至新列
values_to = "score"
) %>%
head(15)更复杂的例子,来自https://epirhandbook.com/en/pivoting-data.html 看看下面这个例子
前面的例子合并的列是相同类型的数据,比如都是数值。但我们也经常见到一些不同类型数据(multiple classes)的列合并,比如下面这个例子
dfmc <-
tribble(
~id, ~obs1_date, ~obs1_status, ~obs2_date, ~obs2_status, ~obs3_date, ~obs3_status,
"A", "2021-04-23", "Healthy", "2021-04-24", "Healthy", "2021-04-25", "Unwell",
"B", "2021-04-23", "Healthy", "2021-04-24", "Healthy", "2021-04-25", "Healthy",
"C", "2021-04-23", "Missing", "2021-04-24", "Healthy", "2021-04-25", "Healthy"
) 我们强行合并一下试试
pivot_longer(dfmc,
cols = -id, #选择要改变的列
names_to = c("index"), #变量名合并至新列
values_to = "record"
)目前这数据肯定还不是tidydata,因为有的数据合并在同一个列里面了。我们可以尝试这样 这里的重点和难点是这个.value
dfmcl <- pivot_longer(dfmc,
cols = -id, #选择要改变的列
names_to = c("obs",".value"), #找到变量里面共同字符串"obs",然后"_"后面的字符作为新变量名(".value"的作用)
names_sep = "_" # 变量分割"_"
)
dfmcl长数据变宽数据,就非常简单了,大家可以看看官方教程 https://tidyr.tidyverse.org/articles/pivot.html?q=.value#wider
留个作业,请把刚刚的dfmcl数据,obs这个列拆分,变成宽数据(wide data)。
5.3.2 拆分列
之前我们曾经看到过这样的数据,将不同的变量合并到了一列。如下,_f和_m分别代表女和男,性别。
read.csv("dataset/heartrate2.csv") 下面我们仍然可以采用tidyr的 separate命令来拆分
heartrate2 <- read.csv("dataset/heartrate2.csv") %>%
separate(patient, into= c("patient", "sex"), sep = "_")
heartrate2